Machine Learning Prototyping with DuckDB and scikit-learn
TL;DR: In this post, we prototype a machine learning workflow using DuckDB for data handling and scikit-learn for modeling.
Introduction
Machine learning prototyping often involves juggling datasets, preprocessing steps, and performance constraints, which can make the process both complex and time-consuming. scikit-learn is one of Python's most popular and powerful libraries for machine learning, offering a large set of utilities for building and evaluating models. In this post, we will explore how DuckDB complements scikit-learn in the model development life cycle, by implementing a penguin species prediction model on Penguins observations in the Palmer Archipelago.
The following implementation is executed in a marimo Python notebook, which is available on GitHub, in our examples repository.
Data Preparation
We start by loading the Palmer Penguins dataset and use DuckDB's COLUMNS(*) expression to filter out any record which contains NA or NULL:
duckdb_conn.read_csv(
"https://blobs.duckdb.org/data/penguins.csv"
).filter(
"columns(*)::text != 'NA'"
).filter(
"columns(*) is not null"
).select(
"*, row_number() over () as observation_id"
).to_table(
"penguins_data"
)
Even if the NA values are removed from the dataset, DuckDB has inferred the column type as VARCHAR at the moment of read_csv. Therefore, we modify the column type to a numeric type, DECIMAL(5, 2), using the ALTER TABLE statement:
duckdb_conn.sql(
"alter table penguins_data alter bill_length_mm set data type decimal(5, 2)"
)
Tip One can also define the schema at import time.
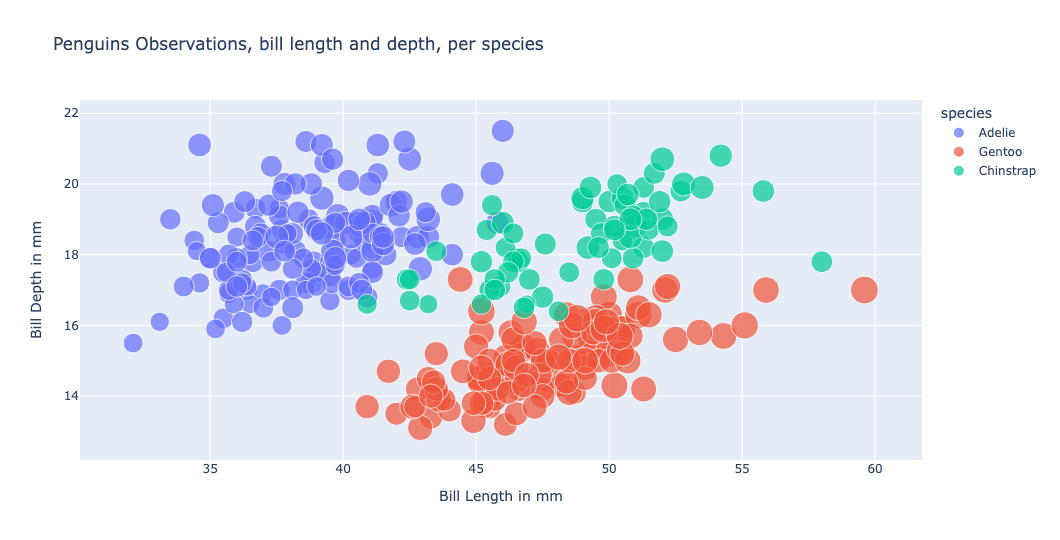
We can now plot the data and check for species specific clusters. For example, using a scatter plot we identify clusters at the combination between bill depth and bill length:
We also observe that there are a few descriptive columns in the dataset, such as species and island. In a machine learning workflow, one common preprocessing step is to transform such values into numerical values, assigning them a unique identifier; a process called label encoding. While scikit-learn offers a LabelEncoder utility, this process is very similar to how we work with reference tables in a data warehouse. Therefore, we define a function which will create, for each column (feature), a reference table, leveraging the DuckDB Python relational API:
def process_reference_data(duckdb_conn):
for feature in ["species", "island"]:
duckdb_conn.sql(f"drop table if exists {feature}_ref")
(
duckdb_conn.table("penguins_data")
.select(feature)
.unique(feature)
.row_number(
window_spec=f"over (order by {feature})",
projected_columns=feature
)
.select(f"{feature}, #2 - 1 as {feature}_id")
.to_table(f"{feature}_ref")
)
duckdb_conn.table(f"{feature}_ref").show()
After the execution of the above function, two tables which will contain the distinct values of the category and a unique identifier are created, e.g., species_ref:
┌───────────┬────────────┐
│ species │ species_id │
│ varchar │ int64 │
├───────────┼────────────┤
│ Adelie │ 0 │
│ Chinstrap │ 1 │
│ Gentoo │ 2 │
└───────────┴────────────┘
The last step in our data preparation step is to define a selection query variable, which selects the data from the initial dataset and the reference tables:
selection_query = (
conn.table("penguins_data")
.join(conn.table("island_ref"), condition="island")
.join(conn.table("species_ref"), condition="species")
)
selection_queryis lazy evaluated, therefore, no data is retrieved at the definition time.
Model Training
Our scope is to predict a penguin's specie based on its characteristics (features), such as bill length and depth, flipper length, body mass and island. Such a model is called a classification model because it predicts the category to which the input data belongs.scikit-learn offers multiple classification models. For our dataset, we have chosen to implement a random forest classifier, based on decision trees:
We start by splitting the data into train and test data, with the train_test_split utility from scikit-learn:
def train_split_data(selection_query):
X_df = selection_query.select("""
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
island_id,
observation_id,
species_id
""").order("observation_id").df()
y_df = [
x[0]
for x in selection_query.order("observation_id").select("species_id").fetchall()
]
num_test = 0.30
return train_test_split(X_df, y_df, test_size=num_test)
X_train, X_test, y_train, y_test = train_split_data(selection_query)
Splitting the data is a common step in a machine learning workflow and it returns:
X_train, containing the input data based on which we want to assign a species category;y_train, containing the species for each record inX_train;X_test, containing the input data based on which we will test the model;y_test, containing the species for each record inX_test.
We then define the RandomForestClassifier and fit the X_train and y_train data to it. We save the model in a pickle file, such that we can use the model without retraining:
model = RandomForestClassifier(n_estimators=1, max_depth=2, random_state=5)
model.fit(X_train.drop(["observation_id", "species_id"], axis=1).values, y_train)
pickle.dump(model, open("./model/penguin_model.sav", "wb"))
We can now check the model accuracy score:
model.score(
X_test.drop(["observation_id", "species_id"], axis=1).values, y_test
)
0.98
We are working with
pickledue to the small size of our model. Other persistence methods are detailed on thescikit-learndocumentation page.
Inference with DuckDB
Inference is the process of using the model to get predictions on (new) data:
model = pickle.load(open("./model/penguin_model.sav", "rb"))
model.predict(...)
When working with DuckDB there are three approaches to retrieve the predicted category from the data. We can use Pandas, or a DuckDB Python UDF with or without batching. In the following, we present these approaches.
With Pandas
The most common method of retrieving predictions is to load the predictions into a new column in a Pandas dataframe:
predicted_df = selection_query.select("""
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
island_id,
observation_id,
species_id
""").df()
predicted_df["predicted_species_id"] = model.predict(
predicted_df.drop(["observation_id", "species_id"], axis=1).values
)
With DuckDB, the dataframe can be queried with SQL:
(
duckdb_conn.table("predicted_df")
.select("observation_id", "species_id", "predicted_species_id")
.filter("species_id != predicted_species_id")
)
resulting in:
┌────────────────┬────────────┬──────────────────────┐
│ observation_id │ species_id │ predicted_species_id │
│ int64 │ int64 │ int64 │
├────────────────┼────────────┼──────────────────────┤
│ 13 │ 1 │ 2 │
│ 15 │ 1 │ 2 │
│ 39 │ 1 │ 2 │
│ 44 │ 1 │ 2 │
│ 68 │ 1 │ 2 │
│ 70 │ 1 │ 2 │
│ 76 │ 1 │ 2 │
│ 90 │ 1 │ 2 │
│ 94 │ 1 │ 2 │
│ 104 │ 1 │ 2 │
│ 106 │ 1 │ 2 │
│ 110 │ 1 │ 2 │
│ 124 │ 1 │ 2 │
│ 126 │ 1 │ 2 │
│ 243 │ 3 │ 2 │
│ 296 │ 2 │ 1 │
├────────────────┴────────────┴──────────────────────┤
│ 16 rows 3 columns │
└────────────────────────────────────────────────────┘
Warning If there is a table which has the same name as the dataframe, then
registercan be used to give another table name to the dataframe, e.g.,duckdb_conn.register("table_name", predicted_df)
DuckDB Python UDF, Row by Row
DuckDB offers the possibility to register User Defined Functions (UDFs) from Python functions. Because the UDF is executed row by row, our prediction function will return the predicted species id for each row:
def get_prediction_per_row(
bill_length_mm: Decimal,
bill_depth_mm: Decimal,
flipper_length_mm: int,
body_mass_g: int,
island_id: int
) -> int:
model = pickle.load(open("./model/penguin_model.sav", "rb"))
return int(
model.predict(
[
[
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
island_id,
]
]
)[0]
)
In the above Python function we provide the required features as input for the model and return the predicted value (an integer). With this information, we create the DuckDB function:
duckdb_conn.create_function(
"predict_species_per_row", get_prediction_per_row, return_type=int
)
We can now use the UDF in the SQL SELECT clause:
selection_query.select("""
observation_id,
species_id,
predict_species_per_row(
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
island_id
) as predicted_species_id
""").filter("species_id != predicted_species_id")
This returns the same results as above.
DuckDB Python UDF, Batch Style
While the row-by-row prediction can be useful when the scope is to retrieve the prediction for a limited number of rows, it will be less performant when working with more data. Therefore, by aggregating the data into arrays (which preserve order), we can simulate a mass retrieval (or batch style) of the prediction.
We first create a Python function to get the predictions for a JSON object, which contains the columnar representation of the features needed as model input:
def get_prediction_per_batch(input_data: dict[str, list[Decimal | int ]]) -> np.ndarray:
"""
input_data example:
{
"bill_length_mm": [40.5],
"bill_depth_mm": [41.5],
"flipper_length_mm: [250],
"body_mass_g": [3000],
"island_id": [1]
}
"""
model = pickle.load(open("./model/penguin_model.sav", "rb"))
input_data_parsed = orjson.loads(input_data)
input_data_converted_to_numpy = np.stack(tuple(input_data_parsed.values()), axis=1)
return model.predict(input_data_converted_to_numpy)
duckdb_conn.create_function(
"predict_species_per_batch",
get_prediction_per_batch,
return_type=duckdb.typing.DuckDBPyType(list[int]),
)
While DuckDB has
MAPandSTRUCTdata types, which convert automatically to a dict, they have a slower execution time compared tojson_object(including theorjsondeserialization time).
With json_object we extract the columnar representation of the features, in the format 'feature_name': array[feature]:
json_object(
'bill_length_mm', array_agg(bill_length_mm),
'bill_depth_mm', array_agg(bill_depth_mm),
'flipper_length_mm', array_agg(flipper_length_mm),
'body_mass_g', array_agg(body_mass_g),
'island_id', array_agg(island_id)
) as input_data,
We then pack in a STRUCT, the predictions together with other columns we are interested in:
struct_pack(
observation_id := array_agg(observation_id),
species_id := array_agg(species_id),
predicted_species_id := predict_species_per_batch(input_data)
) as output_data
Last, we unnest the results, in order to flatten the lists into tables:
.select("""
unnest(output_data.observation_id) as observation_id,
unnest(output_data.species_id) as species_id,
unnest(output_data.predicted_species_id) as predicted_species_id
""")
The above query is wrapped into a Python function, named get_selection_query_for_batch, with which we can chain queries, e.g., for mass retrieval of incorrect predictions:
get_selection_query_for_batch(selection_query).filter("species_id != predicted_species_id")
A batch approach can be implemented by using LIMIT and OFFSET to loop through the data:
for i in range(4):
(
get_selection_query_for_batch(
selection_query
.order("observation_id")
.limit(100, offset=100*i)
.select("*")
)
.filter("species_id != predicted_species_id")
).show()
LIMITandOFFSETare executed last, therefore, they should be applied before the prediction selection.
Performance Considerations
To obtain performance data on a larger scale dataset, we have generated a dummy dataset with approximately 59 million records. On a sample of 10%, on a 16 GB MacBook Pro, the batch processing ranges between 3 and 4 seconds, while the Pandas implementation is executed under 1 second. This is because the Python UDF includes multiple conversion steps which affect performance:
- parsing the input data as JSON;
- converting the JSON to numpy array;
- unnesting the array to rows.
We have still chosen to show-case the Python UDFs, because they are a powerful asset when working within a Python environment and the performance difference is negligible when we speak of small data.
Conclusion
In this post, we have demonstrated how DuckDB complements scikit-learn in the machine learning development life cycle, with a focus on the data preparation and inference phases. The inference results on the dummy data are poor; is this because of the model or the dummy data? We leave this question open to our readers, as a means to explore how DuckDB can be used in the model evaluation and model optimization phases.